Developer Offer
Try ImaginePro API with 50 Free Credits
Build and ship AI-powered visuals with Midjourney, Flux, and more — free credits refresh every month.
Self Learning AI Is Revolutionizing Digital Pathology
The field of digital pathology is on the brink of a major transformation, thanks to the advent of self-learning artificial intelligence. As discussed at the recent Digital Pathology and AI Congress in London, these innovative AI models offer significant advantages over established methods, delivering analyses that are not only faster and more flexible but also completely transparent.
 Image source: Claudio Quiros A, Coudray N, Yeaton A et al., Nature Communications 2024 (CC BY 4.0)
Image source: Claudio Quiros A, Coudray N, Yeaton A et al., Nature Communications 2024 (CC BY 4.0)
Professor John Le Quesne, a leading expert in Molecular Pathology from the University of Glasgow, presented the groundbreaking potential of these approaches, particularly focusing on their application to lung adenocarcinoma and data-rich multiplex images.
 Professor John Le Quesne
Professor John Le Quesne
Leaving the Black Box Behind: Supervised vs Self Learning AI
For years, the development of AI in pathology has relied on classical supervised learning. This method requires expert pathologists to spend countless hours manually annotating images, teaching the AI to recognize specific features. Professor Le Quesne highlighted the drawbacks of this model: "The supervised approach is very expensive in terms of time and salary and runs the danger of ending up with a black box which can give strong opinions but might have more difficulty in explaining how it got there."
In stark contrast, unsupervised self-learning AI represents a paradigm shift. This approach offers several key advantages:
- No Labels Required: The system learns meaningful features directly from the images without any human annotation.
- Speed and Scalability: It is rapid, with its capacity limited only by the available computing power and data.
- Full Interpretability: The results are "wholly interpretable," moving away from the opaque nature of black box models.
How Histomorphological Phenotype Learning Works
Professor Le Quesne focused on a specific self-learning technique called Histomorphological Phenotype Learning (HPL). This method is designed to discover recurring morphological patterns across a large set of histology images and summarize them in a way that is both quantitative and easy to understand. You can learn more about the technical details in the original research paper.
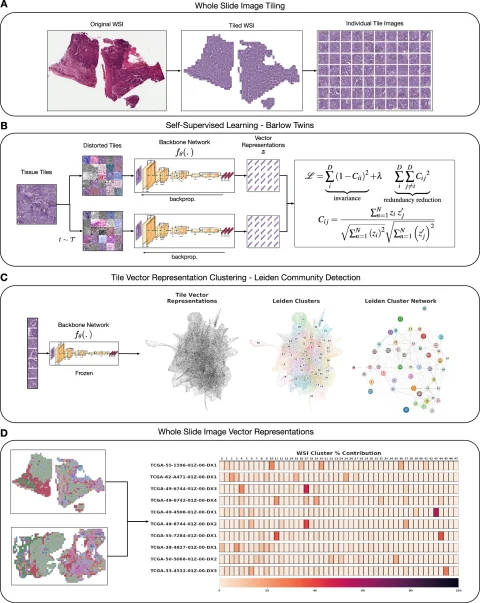 An overview of the HPL framework architecture. Image source: Claudio Quiros A, Coudray N, Yeaton A et al., Nature Communications 2024 (CC BY 4.0)
An overview of the HPL framework architecture. Image source: Claudio Quiros A, Coudray N, Yeaton A et al., Nature Communications 2024 (CC BY 4.0)
The process begins by breaking down whole slide images (WSI) of tissues, such as lung adenocarcinoma, into a vast number of smaller tiles. The HPL system then analyzes these tiles in an unsupervised manner, identifying and grouping tiles with similar morphological features into what are called histomorphological phenotype clusters (HPCs).
This effectively converts a complex H&E image into an intuitive map of these clusters, providing a detailed and quantifiable morphological picture of the tissue. "It gives us a really interpretable quantitative tool to classify images and search tissues," Le Quesne explained. "We can very quickly view the morphological content of that tissue."
A Powerful Tool for Modern Pathology
The practical implications of HPL are profound. By analyzing the percentage composition of different HPCs within a tumor, the model can predict patient outcomes with high accuracy. "HPC composition is highly prognostic in validation cohorts," Le Quesne noted.
He described HPL as a "potent tool" that outperforms not only human pathologists but also traditional supervised AI in crucial tasks like cancer subtyping and prognosis. The key benefits include:
- Automated Identification: It automates the identification of meaningful pathological features.
- Quantitative Dictionary: It provides an interpretable and quantitative morphological "dictionary."
- Superior Performance: It surpasses supervised methods and human experts in key diagnostic and prognostic tasks.
- Discovery of New Features: It has the capability to discover entirely new morphologies relevant to disease biology.
The Road Ahead: Potential and Challenges
The application of self-learning AI is not limited to standard H&E images. Professor Le Quesne highlighted its potential in analyzing other advanced imaging modalities, such as multiplex immunofluorescence (IF), to study the tumor's immune microenvironment.
"With images put through the same process... we find ourselves able to prognosticate outcomes really accurately with greater power than human grading of whole slide images," he said. This opens up exciting possibilities for biomarker discovery and for linking specific biological hallmarks of cancer to real-world patient outcomes, like treatment response.
However, several challenges must be addressed before this technology can be widely implemented. These include refining processes for patient selection, optimizing turnaround times, establishing robust quality control measures, and figuring out how to best integrate these systems into existing laboratory platforms and workflows.
About the Expert
John Le Quesne is a Professor of Molecular Pathology at the University of Glasgow, an Associate Group Leader at the CRUK Research Institute in Glasgow, and an honorary consultant pathologist with NHS Greater Glasgow and Clyde. His research focuses on using quantitative pathological methods to explore human tumor biology.
Reference
Claudio Quiros A, Coudray N, Yeaton A et al.: Mapping the landscape of histomorphological cancer phenotypes using self-supervised learning on unannotated pathology slides; Nature Communications 2024; Read the full paper here.
Compare Plans & Pricing
Find the plan that matches your workload and unlock full access to ImaginePro.
| Plan | Price | Highlights |
|---|---|---|
| Standard | $8 / month |
|
| Premium | $20 / month |
|
Need custom terms? Talk to us to tailor credits, rate limits, or deployment options.
View All Pricing Details